Introduction
This will be just a quick little follow-up to the last post where we created a model and some saved data for returning recipes with similar ingredients. In this post, we'll create an endpoint for the model and return recipes based on the ingredients passed to the FastAPI endpoint.
Setup Script
If you haven't already, please clone the GitHub repository.
git clone https://github.com/kriserickson/recipe-parser.gitThen check out the branch for this blog post.
cd recipe-parser
git checkout unsupervised-1The code is pretty straightforward, so let's go through it section by section.
# Toggle default similarity computation method:
USE_SKLEARN_COSINE = False
PROJECT_DIR = Path(__file__).resolve().parent.parent
MODELS_DIR = PROJECT_DIR / "models"
# ---------- Config: file paths ----------
KMEANS_PATH = MODELS_DIR / "kmeans_model.pkl"
TITLES_PATH = MODELS_DIR / "recipe_titles.pkl"
X_PATH = MODELS_DIR / "tfidf_matrix.joblib"
FILENAMES_PATH = MODELS_DIR / "recipe_filenames.pkl" # <-- new
# ---------- Core search helpers ----------
kmeans = None
titles: List[str] = []
filenames: List[str] = []
X: csr_matrix | None = None
cluster_to_indices: dict[int, np.ndarray] = {}The first section basically just sets up some paths and some variables. The USE_SKLEARN_COSINE variable is a toggle for using sklearn's cosine similarity function or using the dot-product. The KMEANS_PATH variable is the path to the saved kmeans model. The TITLES_PATH variable is the path to the saved titles for the recipes. The X_PATH variable is the path to the saved tfidf matrix. The FILENAMES_PATH variable is the path to the saved filenames for the recipes. The cluster_to_indices variable is a dictionary that maps each cluster to the indices of the recipes in that cluster.
def _build_cluster_index(labels: np.ndarray) -> dict[int, np.ndarray]:
"""Map cluster_id -> np.ndarray of row indices."""
clusters: dict[int, List[int]] = {}
for i, c in enumerate(labels):
clusters.setdefault(int(c), []).append(i)
return {cid: np.asarray(idxs, dtype=np.int32) for cid, idxs in clusters.items()}
@asynccontextmanager
async def lifespan(app):
# startup: load artifacts (same logic as previous _load_artifacts)
global kmeans, titles, filenames, X, cluster_to_indices
try:
with open(KMEANS_PATH, "rb") as f:
kmeans = pickle.load(f)
with open(TITLES_PATH, "rb") as f:
titles = pickle.load(f)
with open(FILENAMES_PATH, "rb") as f:
filenames = pickle.load(f)
X = joblib_load(X_PATH)
if not hasattr(X, "shape"):
raise ValueError("Loaded X is not a sparse matrix.")
# build cluster index from kmeans.labels_
if hasattr(kmeans, "labels_"):
labels = np.asarray(kmeans.labels_, dtype=np.int32)
else:
raise RuntimeError("kmeans.labels_ not found. Refit or save labels separately.")
cluster_to_indices = _build_cluster_index(labels)
# sanity checks
if len(titles) != labels.shape[0]:
raise RuntimeError(
f"titles length ({len(titles)}) != number of samples ({labels.shape[0]}). "
"Artifacts must be built from the same dataset/order."
)
if X is not None and X.shape[0] != labels.shape[0]:
raise RuntimeError(
f"X rows ({X.shape[0]}) != number of samples ({labels.shape[0]}). "
"Artifacts out of sync."
)
if filenames and len(filenames) != labels.shape[0]:
raise RuntimeError(
f"filenames length ({len(filenames)}) != number of samples ({labels.shape[0]}). "
"Save filenames in the same order as titles when building artifacts."
)
yield # application runs after this
finally:
# optional: shutdown cleanup
pass
# create app with lifespan handler
app = FastAPI(title="Recipe Similarity API", version="1.0.0", lifespan=lifespan)The code then sets up the lifespan handler for the app. The lifespan handler is a function that is called when the app starts and stops and is the modern way to handle setting up and tearing down of FastAPI services (the old @app.on_event is now deprecated). In this case, we're loading the saved artifacts from the previous post. We also build the cluster_to_indices dictionary from the kmeans.labels_ attribute. We then do some sanity checks to make sure that the artifacts are built from the same dataset and that the X matrix is the same size as the labels array.
The _build_cluster_index function builds a lookup mapping each cluster id that returns array of row indices that belong to that cluster. It's used so we can quickly retrieve candidate rows for a given cluster instead of scanning all rows.
It works by:
- Create an empty dict clusters:
cluster_id-> list of int. - Iterate enumerate(labels): for each index
iand cluster valuec, appenditoclusters[c](viasetdefault). - Convert each list of indices to a
numpy.ndarray(dtype=np.int32) and return the final dict.
Similar Recipes Endpoint
Let's jump forward in the script and look at the endpoint for similar recipes. Here's the code:
@app.get("/similar_recipes", response_model=SimilarResponse)
def similar_recipes(
recipe_name: str = Query(..., min_length=1),
top_k: int = Query(10, ge=1, le=100), # default to 10 results as requested
fuzzy_cutoff: float = Query(0.35, ge=0.0, le=1.0),
):
"""
It first performs a fuzzy title match. If a good title match is found, use that recipe's
TF-IDF ingredient vector as the query vector and return recipes similar by ingredients.
If no good title match is found, return an empty result set.
"""
# 1) Try fuzzy title match
best_idx, match_score = _best_title_index(recipe_name, cutoff=fuzzy_cutoff)It takes a few options as arguments, but we are mostly interested in the recipe_name argument. The endpoint parameters are defined with FastAPI's Query helper (for example: recipe_name: str = Query(..., min_length=1)). The recipe_name argument is the name of the recipe that we want to find similar recipes for.
We then try to find a good match for the recipe name using the fuzzy title match.
def _best_title_index(name: str, cutoff: float = 0.35) -> tuple[int | None, float]:
"""
Return (best_index, score). Score is in [0..1].
"""
if not titles:
return None, 0.0
best = rf_process.extractOne(
name, titles, scorer=rf_fuzz.partial_token_sort_ratio
)
if best is None:
return None, 0.0
match_val, score, idx = best # rapidfuzz returns (match, score, key)
return int(idx), float(score) / 100.0 The _best_title_index function takes a name. The cutoff value is the minimum score that a match must have to be considered a good match. The function then uses the RapidFuzz extractOne function to find the best match for the name in the list of titles. The scorer in RapidFuzz is a partial token sort ratio which combines partial matching with word reordering which we hope will be good for matching recipe titles. There are a bunch of other scorers available in RapidFuzz, so try experimenting with the various options.
if best_idx is not None and match_score >= fuzzy_cutoff:
query_vector = X[best_idx]
# Determine cluster for that recipe
cluster_id = int(kmeans.labels_[best_idx])
candidate_indices = cluster_to_indices.get(cluster_id, np.array([], dtype=np.int32))
if candidate_indices.size == 0:
candidate_indices = np.arange(X.shape[0], dtype=np.int32)
candidate_indices = candidate_indices[candidate_indices != best_idx]If a good match is found, we find which cluster the index is in (on startup if you remember, we stored the clusters and all the indices into the X array for quick lookup.) Now we have all the indices for cluster we remove the recipe that we found by matching the title (if you are not familiar with python, using a boolean in a NumPy ndarray returns all the elements from that array matching the boolean, under the hood [] calls __getItem__ so it can be easily overloaded as it is in the NumPy ndarray).
if candidate_indices.size == 0:
return SimilarResponse(
query=recipe_name,
cluster=cluster_id,
total_candidates=0,
results=[],
matched_title=titles[best_idx],
matched_filename=(filenames[best_idx] if filenames and filenames[best_idx] else None),
)
candidate_matrix = X[candidate_indices]
top_local, sims = _cosine_similarity_rank(query_vector, candidate_matrix, top_k=min(top_k, candidate_matrix.shape[0]), use_sklearn=USE_SKLEARN_COSINE)Next is a simple sanity to ensure that the cluster exists and wasn't a cluster of 1. Again using the indexing operator on X we grab all the elements in the cluster into the candidate_matrix. Next we call our _cosine_similarity_rank function which ranks the similarities in the cluster our recipe matched.
def _cosine_similarity_rank(query_vector: csr_matrix, candidate_matrix: csr_matrix, top_k: int, use_sklearn: bool ) -> tuple[np.ndarray, np.ndarray]:
"""
Rank candidates by cosine similarity. With TF-IDF default norm='l2',
dot product equals cosine similarity.
Parameters
- q_vec: (1 x D) query sparse row
- cand_matrix:(N x D) candidate sparse matrix
- top_k: number of top results to return
- use_sklearn: if True use sklearn.cosine_similarity (robust, normalizes inside).
if False use sparse dot-product (fast, requires pre-normalized vectors).
Returns (top_local_indices_relative_to_cand_matrix, sims_array)
"""
# timing starts here
start = time.perf_counter()
# Compute similarity scores (1D array length n_cands)
if use_sklearn:
# sklearn will handle normalization and safety checks.
sims = cosine_similarity(query_vector, candidate_matrix).ravel()
method = "sklearn"
else:
# Fast sparse dot-product. Correct only if rows are L2-normalized (TF-IDF default).
sims = (query_vector @ candidate_matrix.T).toarray().ravel()
method = "dot-product"
elapsed = time.perf_counter() - start
logger.info("cosine similarity computed using %s for %d candidates in %.4fs", method, sims.size, elapsed)
n = sims.size
if n == 0:
return np.asarray([], dtype=np.int32), sims
k = min(max(int(top_k), 1), n) # ensure 1 <= k <= n
# np.argpartition is expected to run in linear time O(n) to partition the array and
# place the top-k candidates into the first k positions (unordered). You must
# then fully sort only those k items to produce a correctly ordered top-k list.
# This reduces work compared to sorting all n entries; the benefit grows with
# large n (e.g. hundreds of thousands to millions). For small candidate sets the
# overhead may make the simple argsort approach faster in practice.
part = np.argpartition(-sims, kth=k-1)[:k]
top_local = part[np.argsort(-sims[part])]
return top_local, simsWhile this entire code could be replaced by the much simpler
sims = cosine_similarity(query_vector, candidate_matrix).ravel()
top_local = np.argsort(sims)[::-1][:top_k]
return top_local, simswe have included two (premature because our sample size is so small) optimizations to show how one might optimize both the cosign_similarity ranking of the array, and the sorting of the top_local results. The first optimization we have made optional so that we can log the performance of each algorithm.
The "optimization" is simple: the dot product equals cosine similarity only when both vectors are L2-normalized (i.e. where ||a|| = ||b|| = 1) then the denominator is 1, so cosine(a, b) = a · b).
In scikit-learn, TfidfVectorizer defaults to norm='l2', so the rows produced by vectorizer.fit_transform(texts) are L2-normalized and the dot product between two rows equals their cosine similarity. If you set TfidfVectorizer(norm=None) or perform additional transforms that remove normalization, the dot product no longer equals cosine similarity — use scikit-learn's cosine_similarity (sklearn.metrics.pairwise.cosine_similarity) in that case.
If you compare the two methods you may see the dot-product approach is faster (in our short benchmark it was about twice as fast), but that speedup only matters with large candidate sets. For small datasets the difference is typically negligible:
cosine similarity computed using sklearn for 2656 candidates in 0.0032svs:
cosine similarity computed using dot-product for 2656 candidates in 0.0012showever, if you have a million candidates or more candidates it would be worth the savings.
The second optimization is that instead of just sorting the array and getting the sorted results we use np.argpartion which has the advantage of being linear time for O(n), and then only sorting the top_k values. Since even the fastest sort is O(n log n) this also would mean a considerable savings if the number of candidates was in the millions.
Now that we have our top_k results we just need to format them and display them. Which is done with the _format_results function.
results = _format_results(candidate_indices, sims, top_local)
return SimilarResponse(
query=recipe_name,
cluster=cluster_id,
total_candidates=int(candidate_matrix.shape[0]),
results=results,
matched_title=titles[best_idx],
matched_filename=(filenames[best_idx] if filenames and filenames[best_idx] else None),
)We now have a working endpoint that will take a recipe title and return the recipe it matched as well as some potential similar recipes.
Putting it all together
To view our service in action we probably need a webpage to take in the input, and show the results.
I had CoPilot quickly create a simple front-end for this project which will allow us to test to see how well it works. First, if you haven't already (during the previous article), run the complete unstructured.ipynb code to generate the required models and pickle data files.
We add the following code to our FastAPI service to serve our HTML, JS, and CSS that is in the static directory.
# Serve static SPA
if STATIC_DIR.exists():
app.mount("/static", StaticFiles(directory=str(STATIC_DIR)), name="static")
@app.get("/", include_in_schema=False)
def serve_index():
index_path = STATIC_DIR / "index.html"
if not index_path.exists():
raise HTTPException(status_code=404, detail="index.html not found. Build the SPA under /static.")
return FileResponse(str(index_path))Start the code either with the launch.json entry "Python Debugger: uvicorn similar_service (port 8080) or from the command line with
python -m uvicorn src.similar_service:app --port 8080When you hit the webpage, you will see a prompt to enter a recipe title:
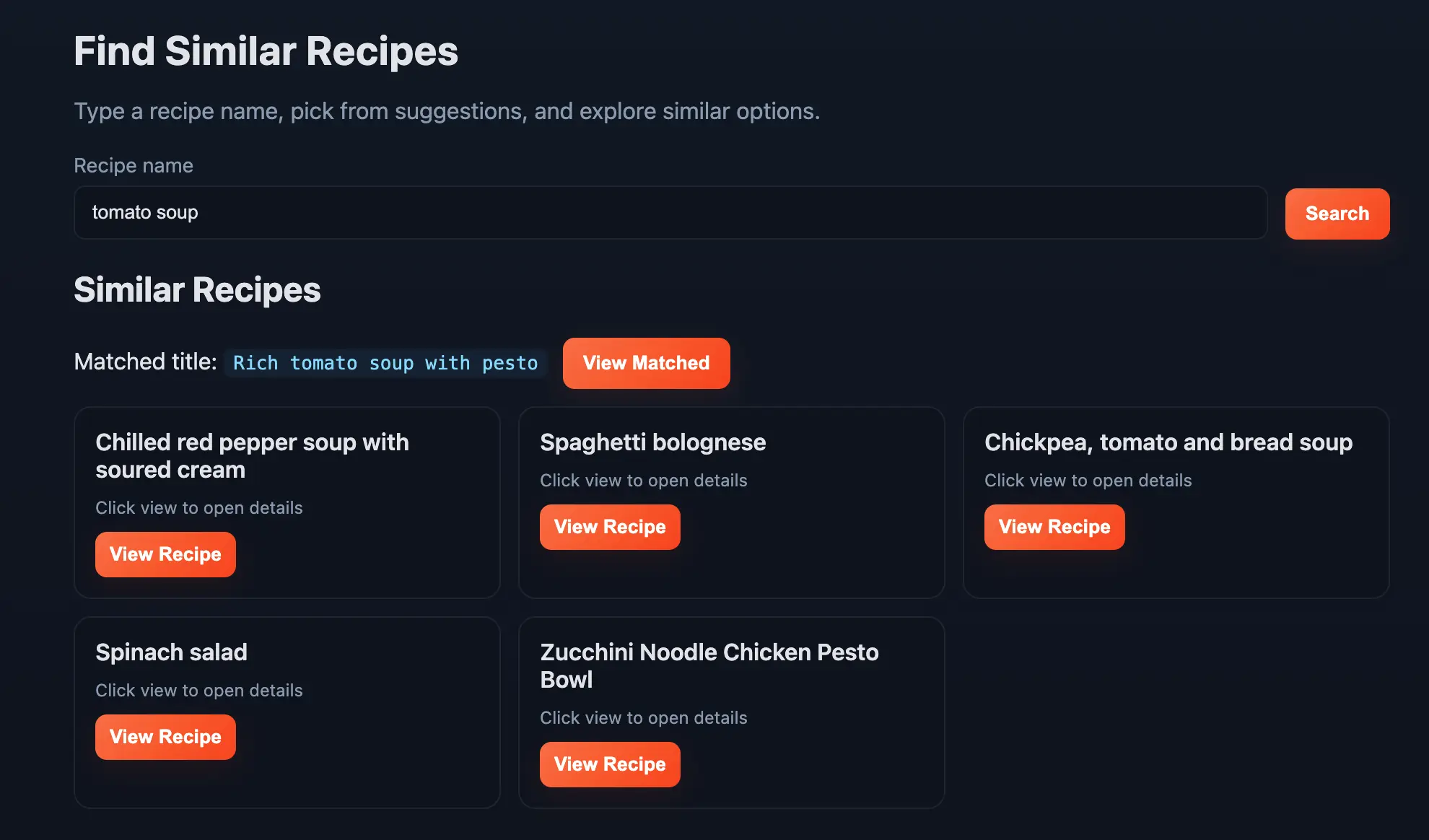
And once you enter a title (like Tomato Soup) you will see some similar recipes:
Final Thoughts
We have quickly created a way of getting similar recipes to a recipe based on its name. So, we come back to the question, why are we clustering these recipes. The goal is to get clusters of recipes that are similar, but not in the exact same way that they are similar in the cosine_similarity of their ingredients. Hopefully the k-means clustering has recognized some similarities in the data not found in the standard similarity of ingredients (some unsupervised learnering as it were). Hopefully, with K-Means, we can discover that recipes tend to fall into, say, “Asian stir-fries,” “Mediterranean salads,” “baked desserts,” etc., based on ingredient patterns and then use the blunt ingredients similarity to bring those to the forefront. The clusters will also speed up the task of finding similar recipes in the end since each cluster is a fraction of the size of the entire corpus of recipes (once again, since we have a small sample this isn't really a benefit).
This may not be the best use of k-means clustering, but it does demonstrate how it works and how it might be used in a real-world situation. We have also learned about cosine_similarity which will show its face again when start looking at RAG over the next few articles. And we can see how quick it is to go from a proof of concept in a Jupyter notebook to a nice REST endpoint (though not a production ready endpoint) and the optimizations that can be made if we understand about what is actually happening with cosine_similarity and inefficient algorithms like sorting.


